RWS-01 The Structure of DNA
 Some parts of this site are currently incomplete & will be updated asap
Some parts of this site are currently incomplete & will be updated asap
Other parts will change continually so use “Refresh” in your browser !!
There is extensive use of “Tooltips” text to support learning which do not seem to render on a Smartphone, so this site is best viewed via a computer’s HD monitor
Learning -> Resources Available, All Work is Subject to DHRF's "GD"
This Research Work Stream follows-on from the previous Research Work Stream
“The structure of DNA is a double helix composed of two intertwined strands” (Hartl 2020, p. 7)
“DNA molecules have two polynucleotides, or “strands,” that wind around an imaginary axis, forming a double helix” (Reece et al. 2014, p. 86)
“In a DNA double helix, two DNA strands are twisted together around a common axis to form a structure that resembles a spiral staircase” (Brooker 2018, p. 219)
“Native DNA Is a Double Helix of Complementary Anti-parallel Strands” (Lodish et al. 2016, p. 170)
“A central feature of double-stranded DNA is complementary base pairing” (Hartl 2020, p. 7)
Discussion in detail
DNA’s different Abstractions
The structure of the DNA polymer can be interpreted in many different ways, depending on the nature of the interest in its structure - these are known as abstractions or models
Various kinds of abstractions (or models) are used all the time, in order to filter out unwanted detail and to show only what is relevant
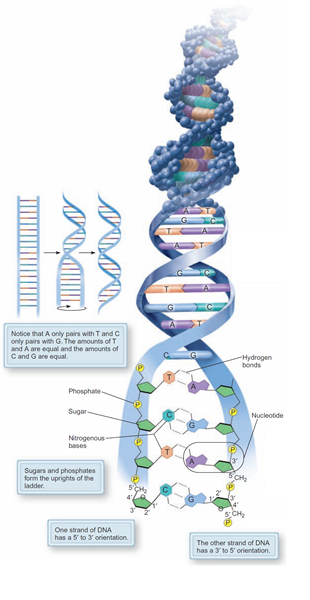
Over on the left is a thumbnail image containing a composite of three different abstractions of the same DNA polymer which illustrates this important concept perfectly
Image: Based upon Fig 8.2 from (Enger et al. 2012, p. 155)
The abstraction that DHRF is concerned with is the DNA structure in the centre which shows the base-pairing of the “rungs” and only then when it has been untwisted
The different types of abstractions are discussed in more detail below
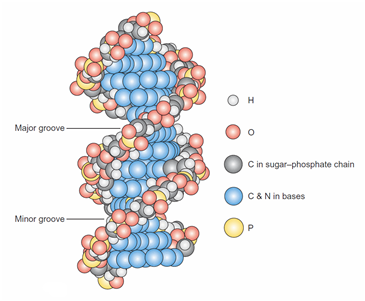
Abstraction#1 - The 3D Molecular View
Fig 2.14(c) from (Weaver 2012, p. 21) is identical to the top section of the composite in (Enger et al. 2012, p. 155) above
“A space-filling model. The sugar–phosphate backbones appear as strings of dark gray, red, light gray, and yellow spheres, whereas the base pairs are rendered as horizontal flat plates composed of blue spheres” (Weaver 2012, p. 21)
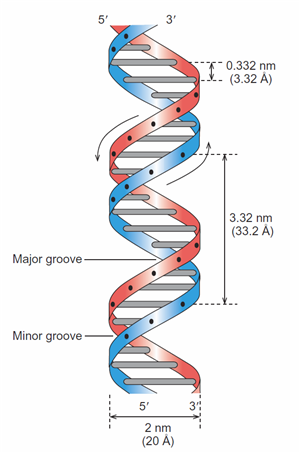
Abstraction#2 - The Double Helix Ladder View
Image: Based upon Fig 2.14(b) from (Weaver 2012, p. 21)
“The DNA double helix is presented as a twisted ladder whose sides represent the sugar–phosphate backbones of the two strands and whose rungs represent base pairs. The curved arrows beside the two strands indicate the 5’ → 3’ orientation of each strand, further illustrating that the two strands are anti-parallel“ (Weaver 2012, p. 21)
This abstraction is concentrating upon the DNA ladder’s physical dimensions
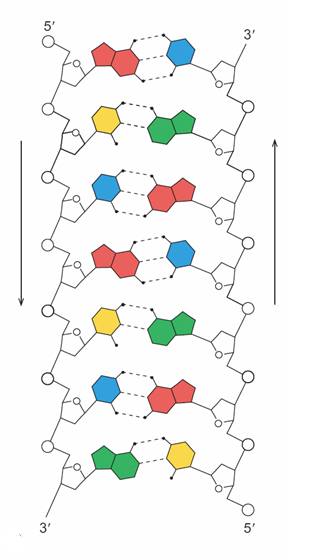
Abstraction#3 - The Chemical Bonding View
Image: Based upon Fig 2.14(a) from (Weaver 2012, p. 21)
“The helix is straightened out to show the base pairing in the middle. Each type of base is represented by a different colour, with the sugar–phosphate backbones in black. Note the three hydrogen bonds in the G–C pairs and the two in the A–T pairs. The vertical arrows beside each strand point in the 5’ → 3’ direction and indicate the anti-parallel nature of the two DNA strands. The left strand runs 5’ → 3’, top to bottom; the right strand runs 5’ → 3’, bottom to top. The deoxyribose rings (white pentagons with O representing oxygen) also show that the two strands have opposite orientations: The rings in the right strand are inverted relative to those in the left strand” (Weaver 2012, p. 21)
Everything so far above is about implementation detail
GD Rule (a)(1)&(2): Ignore implementation detail - focus on functionality
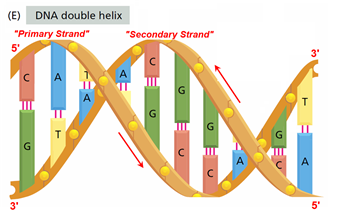
Abstraction#4 - The Double Helix Base Pair (Twisted) View
Image: Based upon Fig 1.2(E) from (Alberts et al. 2008, p. 3) Mark-up in RED by David Husband
This image (Fig 1.2(E) from (Alberts et al. 2008, p. 3) shows the DNA base pairs in the Double Helix
The two base pairs and the four ways in which they are deployed in the Double Helix are of great interest to DHRF. Why?2
![]()
Scientific Method Stage 1 - Make Descriptive Observations:
RWS01/DO-0002 The ‘DNA Double Helix’ is essentially a complex Double Complimentary Linked-List Discussion
![]()
Abstraction#5 - The Base Pair (Untwisted) View
Image: Based upon Fig 1.2(D) from (Alberts et al. 2008, p. 3) Mark-up in RED by David Husband
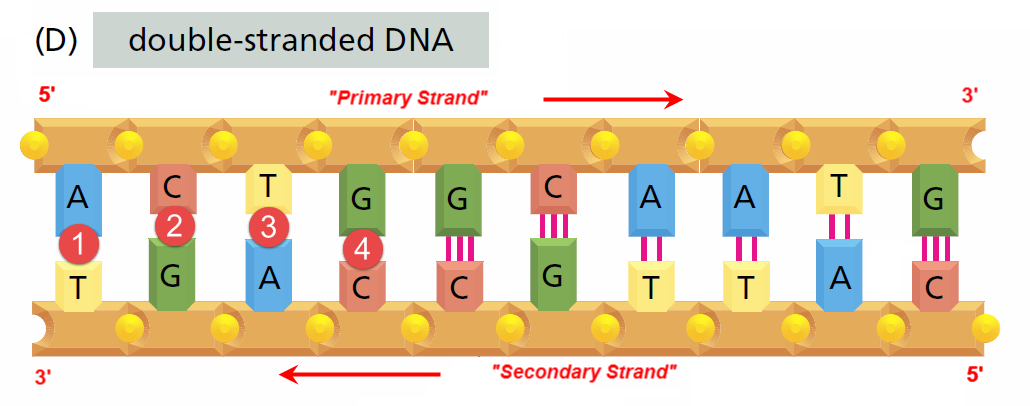
Once the anti-parallel DNA double strand is “unwound” (as above), it can be analysed in more detail
DHRF refers to the Primary Strand GFE-0007 and the secondary Strand GFE-0008 as Functional Equivalences
![]()
Scientific Method Stage 1 - Make Descriptive Observations:
RWS01/DO-0003 The use of complementary base-pairs is a simple parity mechanism Discussion
This is a very simple error-detecting parity mechanism
RWS01/DO-0004 DNA Base Pairs as a 4-Symbol Set
So from the complimentary DNA Base-Pairing rules, A always pairs with T and G always pairs with C suggesting DNA coding involves ONLY TWO SYMBOLS - This is not the case, because:
Refering to Image(D) above, we see, for example, the A-T base-pair in position 1 and the G-C base-pair in position 4, but in position 3 we see an inverted A-T base-pair as a T-A base-pair, and in position 2 we see an inverted G-C base-pair as a C-G base-pair
RWS01/DO-0005 RNA Bases as a 4-Symbol Set Discussion
Scientific Method Stage 4 - Formulate a Hypothesis:
RWS01/HY-0001 DNA has 4 symbols it can code with. ‘DNA-Quad’ is proposed Discussion
![]()
So following on from Observation RWS01/DO-0002 “DO-0003“ above, it is clear that there are 2 DNA Base-pairs, but at the lowest-level, DNA has 4 symbols it can code with
There is already a numeral system called Quaternary that uses the four symbols 0,1,2 & 3
“A quaternary numeral system is base-4. It uses the digits 0, 1, 2 and 3 to represent any real number”
Source: Wikipedia
There are other numeral systems
However, we have already established that DNA uses 4 symbols to code with: A, C, G & T so DHRF proposes a symbol system 3 called DNA-Quad
![]()
Scientific Method Stage 4 - Formulate a Hypothesis:
RWS01/HY-0002 RNA has 4 symbols it can code with. ‘RNA-Quad’ is proposed Discussion
![]()
DHRF has also devised an RNA symbol/numeral system that it calls RNA-Quad
Learning Resources: An Introduction to Number Theory
Abstraction#6 - DHRF’s “Primary Strand” - “Primary Base” View
Image: Based upon Fig 1.2(D) from (Alberts et al. 2008, p. 3) Mark-up in RED by David Husband
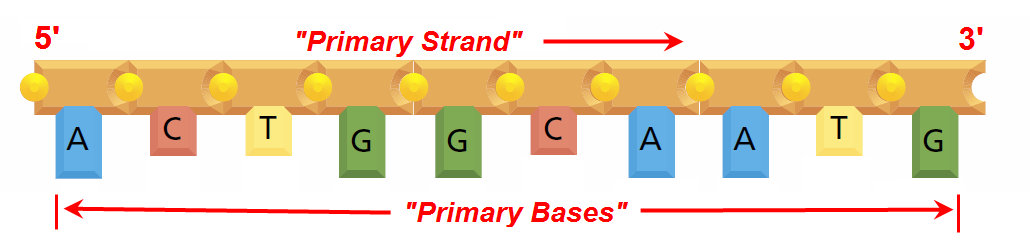
So wrapping up this Research Work Stream, the image above in Abstraction#6 shows the only abstraction DHRF is concerned with as it contains the DNA Sequence of symbols that are used to code the Genome (starting from the left, so in this case the coding sequence is “A C T G G C A A T G“)
Abstraction#7 - DHRF’s “Secondary Strand” - “Secondary Base” View
Image: Based upon Fig 1.2(D) from (Alberts et al. 2008, p. 3) Mark-up in RED by David Husband
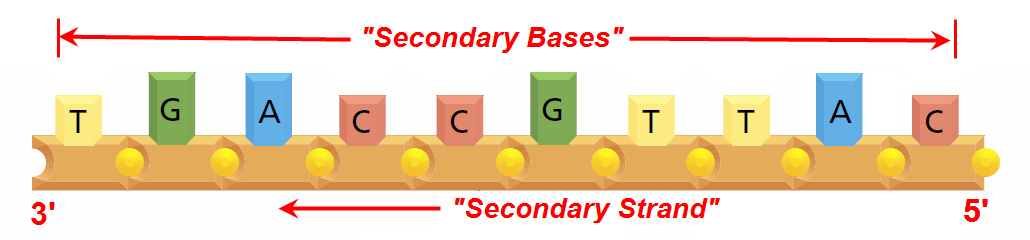
So, finally, for the sake of completeness, DHRF’s “Secondary Strand” - “Secondary Base” abstraction, discussion of which is beyond the scope of this Research Work Stream
Summary - The “Big Picture”…
- DNA makes up an organism’s Genome
- A Chromosome is a long DNA molecule with part or all of the genetic material of an organism
- Genes are contained within chromosomes which contain many different genes
- Codons are part of RNA and specify which amino acid will be added next during protein synthesis, which is a part of “Gene Expression” (“The Central Dogma”)
- The Ribosomes are “macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation)” - Wikipedia
- The Phenotype
This Research Work Stream continues in the next Research Work Stream
References:
Hartl, D. L., 2020. Essential Genetics and Genomics. JONES & BARTLETT PUB INC.
Reece, J. B., Wasserman, S. J., Urry, L. A., Minorsky, P. V., Cain, M. L. and Jackson, R. B., 2014. Campbell Biology. Boston: Pearson.
Lodish, H., Berk, A., Zipursky, S. A., Matsudaira, P., Baltimore, D. and Darnell, J. E., 2016. Molecular cell biology. 8th ed. New York: W.H. Freeman.
Brooker, R., 2018. Genetics: Analysis and Principles. MCGRAW HILL BOOK CO.
Enger, E. D., Ross, F. C. and Bailey, D. B., 2012. Concepts in Biology. 14th ed. New York: McGraw-Hill.
Weaver, R., 2012. Molecular biology. New York: McGraw-Hill.
Alberts, B., Johnson, A., Lewis, L., Raff, M., Roberts, K. and Walter, P., 2008. Molecular Biology of the Cell. 5th ed. New York: Garland Science.
In view of DHRF’s commitment to support learning, there is a much higher “learning content” in the Research Work Streams than would otherwise be the case… Please be aware of that ↩
Because a fundamental understanding of this is required to devise a numeral system that is appropriate for DNA usage. DHRF has therefore devised a DNA symbol/numeral system that it calls “DNA-Quad” and given that RNA has a slightly different base-pair regime, DHRF has also devised an RNA symbol/numeral system that it calls “RNA-Quad”. It is possible in the future that this current proposal will be “tweeked”… ↩
At the moment, all DHRF can establish is that DNA uses a set of 4 symbols (not numbers) to code with at the lowest level ↩